Exercise 3¶
This exercise was generated from a Jupyter notebook. You can download the notebook here.
Problem 3.1: Setting up a Python 2 environment¶
While all major scientific packages are now Python 3 compatible, some are still lagging. It is therefore useful to have a Python 2 distribution available if you want or need it. conda makes this easy! Follow the instructions in Lesson 18 to set up a Python 2 environment.
Problem 3.2: Making your .bashrc file¶
Having a .bashrc file allows you to configure your shell how you like.
a) Create a .bashrc file in your home directory. You can use any text editor you like, even Spyder. If you already have a .bashrc file, open it up for editing.
b) Let's first set the PYTHONPATH environment variable. We didn't explicitly do that in the second command line lesson. The instructions on how to do it are there, though.
c) It is often useful to alias functions to other functions. For example, I am always worried I will accidentally delete things by accident. I therefore have the following in my .bashrc file.
alias rm="rm -i"
Create aliases for commands like ls, mv, and rm based on the flags you like to always use, as you discovered from last tutorial. To figure out what flags are available, you can look at the man pages. If you are using Git Bash on Windows (or if not), asking Google will usually give you the information you need on flags.
If you like, you can download my .bashrc file here.
Problem 3.3: Dropped frames¶
In Lesson 19, we worked on parsing a directory of images from a Leica microscope. The frame rate at which those data were taken was really fast for that microscope. On occasion, the system may miss a frame. Determine if any TIFF files are missing from the directory and, if so, which frame numbers are missing.
Problem 3.4: Installing Biopython, Pysam, and intervaltree¶
In upcoming lessons, we will make use of BioPython, Pysam, and intervaltree. First, install Biopython. It is covered by conda.
conda install biopython
Next, install intervaltree. This uses pip ("Python install Python") for installation. At the command line, do:
pip install intervaltree
Finally, install Pysam using pip. Windows users might have trouble with this. Ask a TA if it's an issue!
Problem 3.5: Generating random sequences¶
It is often useful to generate random DNA and RNA sequences. For example, these are often used to check if an experimentally-observed feature of a sequence is just by chance or not. In this problem, you will generate random sequences under different constraints.
a) Write a function to generate a random sequence with a specified GC content. You should call it like this:
seq = rand_seq_gc(seq_len, gc_content, material='dna')
Here, gc_content can range from zero to one.
b) Write a function to generate a random RNA sequence that can fold into a specified secondary structure. It takes the secondary structure in dot-parentheses format as its single input. As an example,
rand_seq_sec_struc('(((...)))')
could return 'CGCGUAGCG'.
Problem 3.6: Random walks and morphogenesis¶
Diffusion is the process by which a molecule or particle moves randomly in a solution due to constant bombardment by solvent molecules. It can be described as a random walk in which the "walker" randomly takes steps left or right. If the random walker takes a step of length $\delta$ every $\tau$ units of time, the diffusion coefficient is
\begin{align} D = \frac{\delta^2}{2\tau}. \end{align}Molecules are constantly diffusing throughout cells; diffusion is a key process in the transport and organization of molecules.
In the one-cell C. elegans embryo, gradients of morphogens are established on the cell membrane and in the cytoplasm. Specifically, the protein MEX-5 has a gradient in concentration from the anterior to the posterior part of the cytoplasm. An image of this is shown below, taken from Fig. 1 of Griffin, et al., Cell, 146, 955-968, 2011.
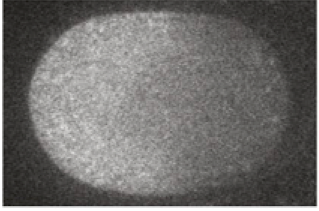
MEX-5 diffuses in the cytoplasm. Griffin and coworkers measured the diffusion coefficient using a FRAP technique. They found that it increased linearly from 1 µm$^2$/s in the anterior (where MEX-5 concentration is observed to be high) to 4 µm$^2$/s in the posterior. The anterior-to-posterior distance is 50 µm.
Based on other observations, Griffin and coworkers speculate that RNA binding to the MEX-5 proteins serves to retard its diffusion. In this problem, we not going to invstigate mechanism, but are interested in whether or not retarded diffusion can account for concentration gradients. To answer this question, we will computationally perform random walks in the cytoplasm in which the step length changes depending on the position within the cell.
a) We will only consider movement along the anterior-posterior axis of the zygote. The anterior is traditionally to the left. So, we model the diffusion as a 1D random walk. First, write a function to take a random walk of n_steps steps starting at position $x_0$ (which is the origin by default). Each step is one unit to the left or to the right. Your function should return the position of the walker at each step as a NumPy array. I.e., your function call should look like this:
walker_positions = unbounded_random_walk(n_steps, x_0=0)
Generate a walk of 10,000 steps. Plot the position of the walker over time. (Here, number of steps is a proxy for time.)
Hint: Unless you are using numba, your code will run much faster if you generate all of your random numbers in one call to one of the functions in the np.random module.
b) Generate 100,000 walks of 1,000 steps each, all starting at the origin. Plot a histogram of the end positions of the walkers.
c) Adjust your unbounded random walk code to take a bounded random walk. Your random walker should proceed as in the unbounded case, but should turn around when it hits a boundary. This would be akin to a molecule diffusing along the anterior-posterior axis of the zygote and hitting the ends of the cell.
Generate a walk of 1000 steps, starting at $x = 25$ on the domain $0 \le x \le 50$, and plot the position of the walker over time.
d) Generate 100,000 walks of 1,000 steps each. (This might take a minute or two to calculate.) This time, each should start at a random position along the anterior-posterior axis, i.e., a random position within the bounds. Plot a histogram of the end positions of the walkers.
Note: I choose the numbers 100,000 walks and 1,000 steps for a reason. Having 100,000 molecules in a C. elegans zygote amounts to a concentration of tens of nanomolar, a reaonsable physiological concentration. We we take the diffusion coefficient to be 1 µm$^2$/s, we can take steps of size $\delta = 1$ µm with $\tau = 1$ second. The time scale of the formation of the MEX-5 gradient is about 1000 seconds, or about 15 minutes. The actual "steps" due to bombardment of the molecule with solvent are far shorter in distance and time, but, as we can derive from the fundamental physics, we can model the random walk as longer steps with a longer time interval.
e) Finally, adjust your unbounded random walk code so that the diffusion coefficient varies linearly from 1 µm$^2$/s at the anterior to 4 µm$^2$/s at the posterior. Perform and plot random walk as in part (c).
f) Generate 100,000 walks of 1,000 steps each and make plots as in part(d). Is the changing diffusion coefficient hypothesis sufficient to explain the presence of a MEX-5 gradient?