Lesson 38: Interval trees and efficient genomic queries¶
This tutorial was generated from a Jupyter notebook. You can download the notebook here.
A common application when manipulating genomic data are queries on genomic intervals. For example, trying to find all genes within a given genomic region or identifying other transcript isoforms that overlap a gene of interest.
The naive approach of simply storing all features and searching for it through iteration can be very inefficient especially for large datasets (such as those that are common in genomics).
Because of the nature of genomic intervals, there are more efficient data structures that enable faster queries based on position.
The approach used is based on a general class of solutions and datastructures known as an Interval Tree
There is an implementation of an Interval Tree in python that can be installed as follows:
pip install intervaltreeNow that we have this installed, let's explore the functionality a bit.
import intervaltree
First, let's build a tree from a set of genes:
tree = intervaltree.IntervalTree() # Initialize an empty tree
with open('chr3.genes.bed', 'r') as f:
for line in f:
tokens=line.split("\t")
tree.addi(int(tokens[1]), int(tokens[2]), tokens[3])
We now have a tree that contains the positions of all genes on mouse chromosome 3
Let's find all genes at a specific location. We'll use the Sox2 locus, which has these 2 genes
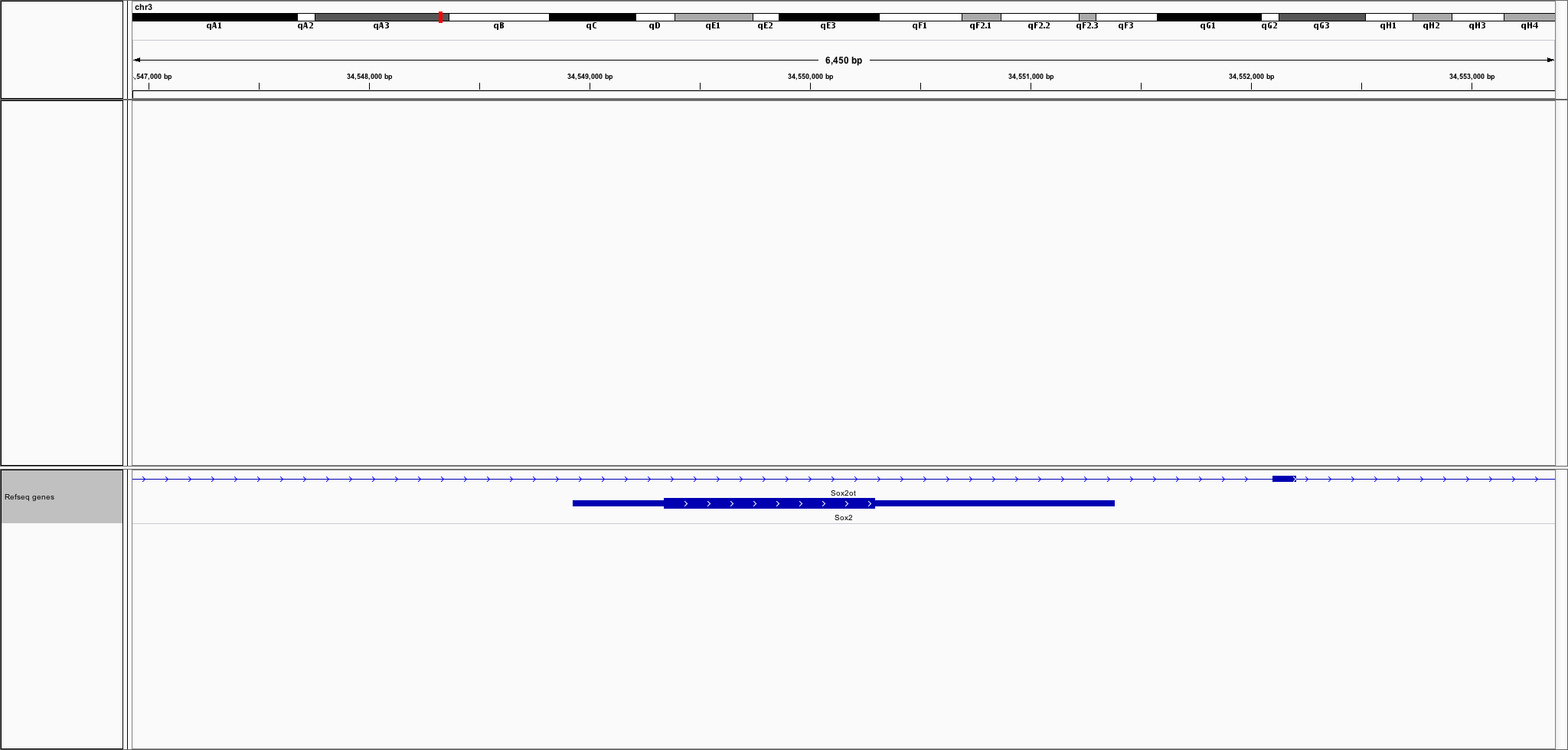
tree.search(34548927, 34551382)
But, you can even look at larger regions such as the following one with many genes:
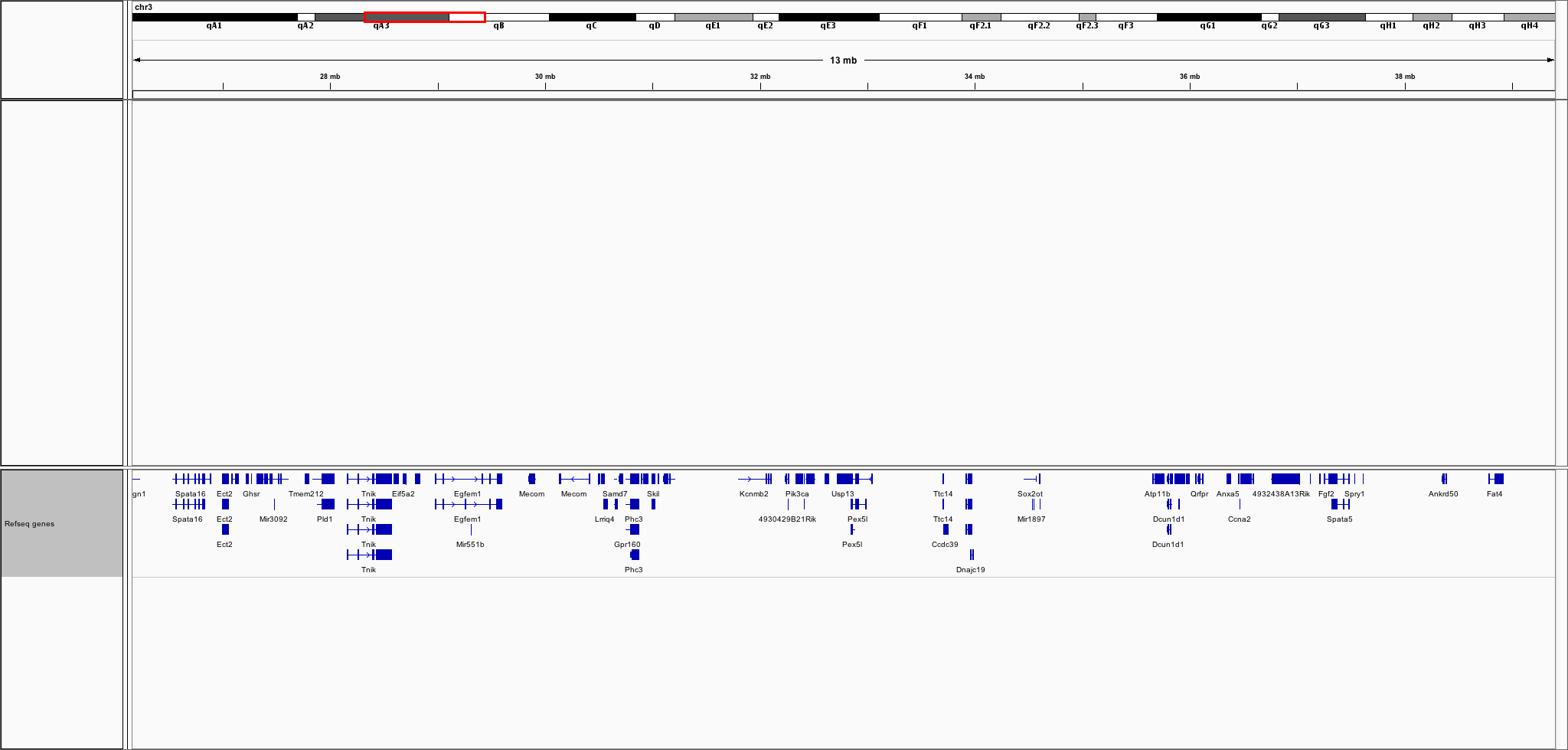
tree.search(26156916, 39409396)
The return type is a set and so any standard iteration methods will work for going through the list.